| PDB Features | |||||||||||||||||||||||||||||||
| PDB ID |
ID of the original structure extracted from the Protein Database (PDB). The search is case insensitive.
Use comma (,) or spaces to specify a list of ID's. Example: 3cvt,1SFU 1m3Q 1b72 Copy to form |
||||||||||||||||||||||||||||||
| Pubmed ID |
ID of the PubMed reference that mentions the structure(s) of interest. Use comma (,) or spaces to specify a
list of ID's. Example: 9367757 11427887, 14739929 Copy to form |
||||||||||||||||||||||||||||||
| Resolution |
Resolution in Angstroms (Å) of the X-Ray structure. |
||||||||||||||||||||||||||||||
| Species (Source) |
The source species of the structure. Select an available species from the list. |
||||||||||||||||||||||||||||||
| No. Complexes per PDB |
The number of independent complexes that appear in a PDB structure (asymmetric unit). Sometimes this number
matches the number of biological units in the X-ray crystal. For example, the structure with PDB ID
1am9 has two complexes,
while the structure with PDB ID 1h89
has only one complex. Select an available number from the list. |
||||||||||||||||||||||||||||||
| No. Interfaces per Complex |
The number of protein-DNA interfaces per complex. An interface is defined when one or more protein subunits
interacting with DNA can be isolated. For example, the structure with PDB ID
1am9 has two complexes each with
one interface (a dimer interacting with DNA) while the structure with PDB ID
1h89 has one complex with two
interfaces (one with a monomer interacting with DNA and one with a dimer interacting with DNA). Select an available number from the list. |
||||||||||||||||||||||||||||||
| Asymmetric Unit = Biological Unit |
Select those stuctures where the Asymmetric Unit of the PDB matches (does not match) its Biological Unit.
|
||||||||||||||||||||||||||||||
| No. Biological Units |
The number of biological units the original structure has according to the information availbale in PDB. Sometimes this number
matches the number of complexes in the X-ray crystal. For example, according to the information available in PDB the
structure with ID 1am9 has one biological unit,
whilst the structure with PDB ID 3f21
has five biological units. Select an available number from the list. |
||||||||||||||||||||||||||||||
| Water Molecules |
Select those stuctures that has (not) got water molecules.
|
||||||||||||||||||||||||||||||
| Protein Features | |||||||||||||||||||||||||||||||
| Class | Function-based classification of the complex from the point of view of the protein part. There are four categories: Enzyme (if the main function of the protein is to modify DNA), Transcription factor (if the main function of the protein is to regulate transcription and gene expression), Structural/DNA Binding protein (if the main function of the protein is to support DNA structure, bend DNA or aggregate other proteins), and Immunological protein (if the DNA/protein interaction triggers response of the immune system). | ||||||||||||||||||||||||||||||
| Type | Function/structure-based classification. This classification uses as a source of information PubMed, PDB, CATH and SCOP. There are 19 types for Enzyme (Methyltransferase, Repair Protein, Topoisomerase, Nuclease, Recombinase, Glucosyltransferase, Transposase, Phosphodiesterase, Excisionase, Kinase, Photolyase, Helicase, Ligase, Translocase, Helicase, Endonuclease, Polymerase, Glycosylase); 8 types for Transcription factor (Zinc Coordinating, Zipper Type, Alpha/Beta, Alpha Helix, Ribbon/Helix/Helix, Beta Sheet, Helix Turn Helix); 8 types for Structural/DNA Binding protein (Replication, Maintenance/Protection, Recombination, Zalpha, Centromeric Protein, Telomeric Protein, Structural Protein); and one type for Immunological protein (Immunoglobulin). | ||||||||||||||||||||||||||||||
| Subtype |
Classification that concerns a more specific type. This classification takes into account e.g. domains, specific reaction of an enzyme,
specific DNA binding sites, etc. Example: Homeodomain Copy to form |
||||||||||||||||||||||||||||||
| No. Protein Monomers per Interface |
The number of protein monomers taking part in the interface (protein interacting with DNA). Select an available number from the list. |
||||||||||||||||||||||||||||||
| Multimerization |
Multimerization of the protein at the interface. There are four possible categories:
|
||||||||||||||||||||||||||||||
| Protein-Protein interaction |
This field represents the way multimeric proteins are interacting in the interface. There are four possible values:
|
||||||||||||||||||||||||||||||
| DNA Features | |||||||||||||||||||||||||||||||
| Double/Single Strand |
There are five possible values:
|
||||||||||||||||||||||||||||||
| Sticky Ends |
Bases that are unpaired at the ends of the double stranded DNA. There are five possible values:
|
||||||||||||||||||||||||||||||
| Flipped Base(s)) |
Flipped bases are bases that does not form Watson-Crick H-bonds inside the molecule.
|
||||||||||||||||||||||||||||||
| Nicked DNA |
Nicked DNA is a DNA molecule with a broken PO4-OH bond.
|
||||||||||||||||||||||||||||||
| Gapped DNA |
Gapped DNA is a DNA molecule that lacks one or more bases in the middle of one strand.
|
||||||||||||||||||||||||||||||
| Open DNA |
Open DNA is a DNA that has bases not forming Watson-Crick H-bonds at the ends of the molecule.
|
||||||||||||||||||||||||||||||
| Modified DNA |
Modified DNA is a DNA molecule that contains chemically modified or not standard bases.
|
||||||||||||||||||||||||||||||
| Cruciform |
Cruciform donotes a DNA molecule that takes part in a cruciform.
|
||||||||||||||||||||||||||||||
| Z-DNA |
Z-DNA is a DNA molecule in Z conformation (left-handed helix).
|
||||||||||||||||||||||||||||||
| Interface Features | |||||||||||||||||||||||||||||||
| Protein-DNA Interaction |
This field represents the way the protein is interacting with the DNA in the interface. There are three possible values:
|
||||||||||||||||||||||||||||||
| Sequence Clustering Group |
Several sequence groups were obtained by aligning the protein sequences (chains) that interact with DNA in a pairwise fashion. Sequences were clustered in groups according to a minimal length coverage threshold of 90% and percentage sequence identity of 70%. This means that two interfaces belong to the same group if any two protein sequences from both interfaces have 70% of the residues identical in at least 90% of the length of both sequences. Because the clustering was done at the protein chain level, a given interface may belong to two or more groups.
|
||||||||||||||||||||||||||||||
| Interface Clustering Group |
Clustering of effective atomic interfaces between DNA and proteins. To perform a clustering of protein-DNA complexes based on their effective distance-dependent matrices, we calculated the following dissimilarity measure (DM) between two matrices Ma and Mb 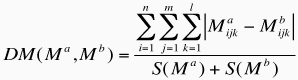 where S(x) is the total number of effective interactions recorded for a given complex, and corresponds to: 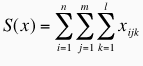 n is the number of protein atom types (40), m is the number of DNA atom types (26) and l the number of distance classes (5). DM values are in the range [0,1], where 0 means that both matrices are identical and 1 means that both complexes have no effective interactions in common. The DM was computed for all pairs of DNA-protein complexes, a difference table built and hierarchical clustering carried out with the group average algorithm. The cutoff used in this case was 0.25 to define the groups. This means that two interfaces are clustered together if they have more than 75% of their effective interactions in common. |
||||||||||||||||||||||||||||||
| No. Effective Contacts |
Total number of effective contacts in the protein-DNA interface.
|
||||||||||||||||||||||||||||||
| Groove Contacts |
Percentages of effective contacts with DNA atoms according to the position in the grooves. Atoms belonging to a specific groove location were
assigned according to the classical definition in B-DNA. So there are atoms in the Major Groove, Minor Groove, Backbone (phosphates and
sugar) and atoms assigned to any location (i.e. ambiguous position).
|
||||||||||||||||||||||||||||||
| Interaction Classes/Types |
Percentages of effective contacts matching the definition of interaction classes/types according to atom identity:
|
||||||||||||||||||||||||||||||