|
 |
 |
 |
 |
|
|
| |
  |
|
 This
is a server that calculates the annealing or melting temperature
of any given short DNA sequence (in the range of 16-30 nts)
using five different approximations. A merged or consensus
temperature among all calculations is also given. In addition
to this, the server will inform to the user about the expected
variation of the melting temperature estimation, which depends
on the specific oligonucleotide sequence. In a recent study
(Panjkovich and Melo, 2005), we have demonstrated
that large variations in the melting temperature estimations
are observed among different methods. The magnitude of the
observed differences depends on the length and CG-content
of the oligonucleotide sequence, but it cannot be explained
in a trivial manner. In fact, complex relationships are observed:
in some cases two or more methods give similar melting temperature
values (within 5° C) for a significant fraction (greater
than 80%) of a large set of oligonucleotides (2000 different
sequences) of fixed length and similar CG-content, but it
turns out that these values are finally highly uncorrelated. This
is a server that calculates the annealing or melting temperature
of any given short DNA sequence (in the range of 16-30 nts)
using five different approximations. A merged or consensus
temperature among all calculations is also given. In addition
to this, the server will inform to the user about the expected
variation of the melting temperature estimation, which depends
on the specific oligonucleotide sequence. In a recent study
(Panjkovich and Melo, 2005), we have demonstrated
that large variations in the melting temperature estimations
are observed among different methods. The magnitude of the
observed differences depends on the length and CG-content
of the oligonucleotide sequence, but it cannot be explained
in a trivial manner. In fact, complex relationships are observed:
in some cases two or more methods give similar melting temperature
values (within 5° C) for a significant fraction (greater
than 80%) of a large set of oligonucleotides (2000 different
sequences) of fixed length and similar CG-content, but it
turns out that these values are finally highly uncorrelated.
 Based
on these results, we suggest that additional experimental
data is required in order to eliminate the existing bias of
the current methods and parameters towards oligonucleotide length and composition
(most of the methods have been parameterized based on a very
restricted set of oligos for which experimental data is currently
available). Meanwhile, the best solution to avoid a large
error in the melting temperature estimation of a given oligonucleotide
sequence is to attempt to get a consensus value among the
existing methods that consistently show similar values for
a given length and CG content. This is what this server attempts
to do, based on the large scale comparative study that we
have performed. Accurate melting temperature estimation could
not be highly relevant for a classic single-locus PCR, but
it becomes extremely important in other applications such
as multiplex PCR, quantitative PCR and the design of fixed
short length oligonucleotide microarrays (Affymetrix-like
chips), where dozens or thousands of DNA molecules are used
simultaneously under identical experimental conditions and,
of course, at a single and fixed temperature. Based
on these results, we suggest that additional experimental
data is required in order to eliminate the existing bias of
the current methods and parameters towards oligonucleotide length and composition
(most of the methods have been parameterized based on a very
restricted set of oligos for which experimental data is currently
available). Meanwhile, the best solution to avoid a large
error in the melting temperature estimation of a given oligonucleotide
sequence is to attempt to get a consensus value among the
existing methods that consistently show similar values for
a given length and CG content. This is what this server attempts
to do, based on the large scale comparative study that we
have performed. Accurate melting temperature estimation could
not be highly relevant for a classic single-locus PCR, but
it becomes extremely important in other applications such
as multiplex PCR, quantitative PCR and the design of fixed
short length oligonucleotide microarrays (Affymetrix-like
chips), where dozens or thousands of DNA molecules are used
simultaneously under identical experimental conditions and,
of course, at a single and fixed temperature.
 The calculations performed in
the consensus Tm estimation method are described in the Figure below. For details about
the thermodynamic calculations see the Methods section. of
supplemental material. The calculations performed in
the consensus Tm estimation method are described in the Figure below. For details about
the thermodynamic calculations see the Methods section. of
supplemental material.
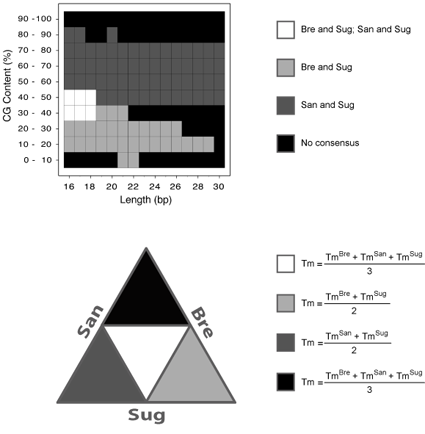
(Top panel) The consensus map from the previous comparative benchmark (Panjkovich and Melo, 2005)
is illustrated. In this benchmark, three thermodynamic data sets were compared: Bre stands for
Breslauer et al. 1986; San stands for SantaLucia et al. 1996; Sug stands for Sugimoto et al. 1996.
In this map, four distinct regions were obtained: 1) simultaneously, Bre and Sug on the one hand,
and San and Sug on the other, exhibited similar Tm values (white colour); 2) only Bre and Sug
exhibited similar Tm values (light gray colour); 3) only San and Sug exhibited similar Tm values
(dark gray colour); and finally, 4) no consensus was observed among any of the methods (black colour).
Bre and San did not show a similar behavior in the complete range of sequence length and percentage
of CG-content. (Bottom panel) A graphical illustration of the different consensus map zones is shown.
Each method is represented as a particular side of an equilateral triangle and the intersection among
methods is shown with the corresponding color of the consensus map. The mathematical expressions used
to calculate the consensus Tm at each zone are also indicated. In the case of San calculations the
most recent thermodynamic parameters (SantaLucia, 1998) are being used by the server to calculate
the consensus melting temperature. This modification respect to our previous study (Melo and Panjkovich, 2005)
has improved even more the accuracy of this server. The Tm estimations of oligonucleotides falling into
the black regions of the consensus map by any of the methods could have a large error. The Tm estimation
error at the other regions where some consensus was observed is expected to be small (below 3-5 ºC).
|
  |
 There
are three different methods that can be used to calculate
the melting temperature of short DNA sequences. The first
one is called the 'Basic' method (bas),
which estimates the melting temperature only based on the
composition of nucleotides. The second method is called 'Salt
Adjusted' (sal),
which is similar to the previous method but includes a logarithmic
factor to correct for salt concentration. The third method
is called the 'Thermodynamic' method, which
uses the nearest-neighbor model and the experimental values
of enthalpy and entropy to get the free energy of duplex formation.
This method also includes additional terms to correct the
melting temperature estimation for the effects of oligo and
salt concentration. There
are three different methods that can be used to calculate
the melting temperature of short DNA sequences. The first
one is called the 'Basic' method (bas),
which estimates the melting temperature only based on the
composition of nucleotides. The second method is called 'Salt
Adjusted' (sal),
which is similar to the previous method but includes a logarithmic
factor to correct for salt concentration. The third method
is called the 'Thermodynamic' method, which
uses the nearest-neighbor model and the experimental values
of enthalpy and entropy to get the free energy of duplex formation.
This method also includes additional terms to correct the
melting temperature estimation for the effects of oligo and
salt concentration.
 Currently,
there are three thermodynamic tables that have been published
in the literature and are widely used for primer melting temperature
calculation using the nearest neighbor approach. These are
the Breslauer (Breslauer et al., 1986) table (defined
as Th1),
the Santalucia (Santalucia, 1998) table (defined
as Th2),
and the Sugimoto (Sugimoto et al., 1996) table (defined
as Th3).
This server will calculate the melting temperature of one
or more oligonucleotides by using the three different methods
and these existing tables of thermodynamic parameters mentioned above, thus giving five different melting temperature
values. In addition to this, a consensus or merged annealing
temperature based on the three thermodynamic sets will be also given, which is expected to have
the smaller error in the long run (average error of several
independent estimations). A detailed description of the mathematical
expressions and experimental data tables used to calculate
the melting temperature of each method is provided here. Currently,
there are three thermodynamic tables that have been published
in the literature and are widely used for primer melting temperature
calculation using the nearest neighbor approach. These are
the Breslauer (Breslauer et al., 1986) table (defined
as Th1),
the Santalucia (Santalucia, 1998) table (defined
as Th2),
and the Sugimoto (Sugimoto et al., 1996) table (defined
as Th3).
This server will calculate the melting temperature of one
or more oligonucleotides by using the three different methods
and these existing tables of thermodynamic parameters mentioned above, thus giving five different melting temperature
values. In addition to this, a consensus or merged annealing
temperature based on the three thermodynamic sets will be also given, which is expected to have
the smaller error in the long run (average error of several
independent estimations). A detailed description of the mathematical
expressions and experimental data tables used to calculate
the melting temperature of each method is provided here.
|
  |
 For
detailed information about the comparative study we have carried
out, read the following paper: Panjkovich and Melo (2005).
For additional data, check out the following link containing
some supplementary material. For
detailed information about the comparative study we have carried
out, read the following paper: Panjkovich and Melo (2005).
For additional data, check out the following link containing
some supplementary material.
|
  |
 Panjkovich,
A., Norambuena, T. and Melo, F. (2005) dnaMATE: a consensus
melting temperature prediction server for short DNA sequences.
Nucleic Acids Research, 33, 570-572. Panjkovich,
A., Norambuena, T. and Melo, F. (2005) dnaMATE: a consensus
melting temperature prediction server for short DNA sequences.
Nucleic Acids Research, 33, 570-572.
 Panjkovich,
A. and Melo, F. (2005) Comparison of DNA melting temperature
calculation methods for short DNA sequences. Bioinformatics
21, 711-722. Panjkovich,
A. and Melo, F. (2005) Comparison of DNA melting temperature
calculation methods for short DNA sequences. Bioinformatics
21, 711-722.
 Breslauer,
K.J., Frank,R., Blöcker, H. and Marky, L.A. (1986) Predicting
DNA Duplex stability from the base sequence. Proc.
Natl. Acad. Sci. USA 83, 3746-3750. Breslauer,
K.J., Frank,R., Blöcker, H. and Marky, L.A. (1986) Predicting
DNA Duplex stability from the base sequence. Proc.
Natl. Acad. Sci. USA 83, 3746-3750.
 SantaLucia
, Jr. J., Allawi, H.T. and Seneviratne, P.A. (1996) Improved Nearest-Neighbor Parameters for
Predicting DNA Duplex Stability.Biochemistry 35, 3555-3562. SantaLucia
, Jr. J., Allawi, H.T. and Seneviratne, P.A. (1996) Improved Nearest-Neighbor Parameters for
Predicting DNA Duplex Stability.Biochemistry 35, 3555-3562.
 SantaLucia, Jr. J. (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics.
Proc. Natl. Acad. Sci. USA 95, 1460-1465. SantaLucia, Jr. J. (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics.
Proc. Natl. Acad. Sci. USA 95, 1460-1465.
 Sugimoto
, N., Nakano, S., Yoneyama, M. and Honda, K. (1996) Improved
thermodynamic parameters and helix initiation factor to predict
stability of DNA duplexes.Nucleic Acids Res. 24, 4501-4505. Sugimoto
, N., Nakano, S., Yoneyama, M. and Honda, K. (1996) Improved
thermodynamic parameters and helix initiation factor to predict
stability of DNA duplexes.Nucleic Acids Res. 24, 4501-4505.
|
|
|