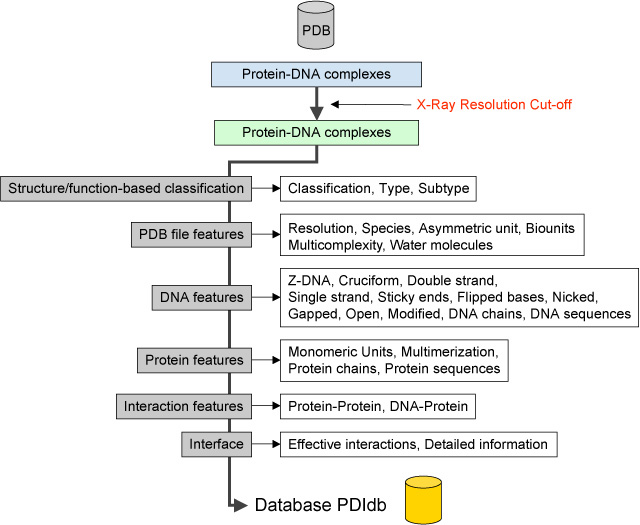
PDIdb (Protein-DNA Interface database) is a repository containing relevant structural information of Protein-DNA complexes solved by X-Ray cristallography and available at the Protein Data Bank (PDB). The database includes a simple functional classification of the protein-DNA complexes that consists of three hierarchical levels (Class, Type and Subtype). This classification has been defined and manually curated by humans based on the information gathered from several sources that included PDB, PubMed, CATH and SCOP. The current version of the database contains only structures with resolution of 2.5 Å or higher. A total of 922 entries are currently available. A detailed flowchart of the process used to build this database is shown at the bottom of this page.
The major aim of this database is to contribute to the understanding of the main rules that underlie the molecular recognition process between DNA and proteins. To this end, we have focused on each specific atomic interface rather than on the separated binding partners (e.g. protein or DNA molecules). Therefore, each entry in this database consists of a single and independent protein-DNA interface.
To remove similar protein-DNA complexes in this database, we have also used two different clustering/grouping schemes of the data: 1) a clustering based on protein sequence identity and fraction of aligned regions for those proteins that form part of the interface with DNA; and 2) a clustering based on the effective interactions observed for each interface between proteins and DNA.
The sequence-based clustering was obtained by aligning the protein sequences (chains) that interact with DNA in a pairwise fashion. Sequences were clustered in groups according to a length coverage threshold of 90% and percentage sequence identity of 70%, using blastclust. Therefore, two protein-DNA interfaces are clustered together if any two protein chains found at the two interfaces share more than 70% sequence identity for at least 90% of the length of both sequences. A total of 246 representative interfaces, out of the initial 922 entries, are obtained with this procedure. The detail of the representative members of the groups can be accessed through this query. Additionaly, a set of PDB files of the representative members as well as other related sets can be downloaded here.
On the other hand, to perform a clustering of protein-DNA complexes based on their effective distance-dependent matrices, we calculated the following dissimilarity measure (DM) between two matrices Ma and Mb
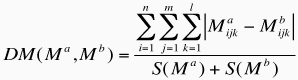
where S(x) is the total number of effective interactions recorded for a given complex, and corresponds to:
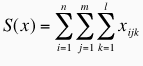
n is the number of protein atom types (40 types), m is the number of DNA atom types (26 types) and l the number of distance classes (5 classes of range 1.4 Å). DM values are in the range [0,1], where 0 means that both matrices are identical and 1 means that both complexes have no effective interactions in common. The DM was computed for all pairs of DNA-protein complexes, a difference table built and hierarchical clustering carried out with the group average algorithm. The cutoff used in this case was 0.25 to define the groups. This means that two interfaces are clustered together if they have more than 75% of their effective interactions in common. A total of 671 representative interfaces, out of the initial 922 entries, are obtained with this procedure.
The detail of the representative members of the groups can be accessed through this query. Additionaly, a set of PDB files of the representative members as well as the cluster tree in postscript format can be found here.
In the near future a new third clustering/grouping scheme, that will be based on the three-dimensional shape and atomic nature of the interfaces, will be incorporated.
We hope that this database will be useful to those people working in the fields of prediction of transcription factor binding sites in DNA, study of specificity determinants that mediates different enzyme recognition events, engineering and design of new transcription factors with distinct binding specificity and affinity, etc. Due to its friendly and easy-to-use web interface, we hope that this database will also serve educational and teaching purposes.