PDIviz

Visualization and Analysis of Protein-DNA Binding Interfaces
Manual: PDIviz guide
Download: PDIviz124a
Version: PDIviz 1.2.4a [2022-10-03]
Changes:
- v1.2.4a [2022-10-03] Added support for PyMOL 2. Tested on PyMOL 2.5.2 Incentive Product. Known issues: Data export does not work.
- v1.2.3 [2018-03-12] Added support for old PDB formats
Authors: Judemir Ribeiro, Francisco Melo, Andreas Schüller
Contact: aschueller@bio.puc.cl (aschueller null@null bio NULL.puc NULL.cl)
Licensed under the terms of the MIT license. Copyright (c) 2015 Judemir Ribeiro, Francisco Melo, Andreas Schüller
Incorporates the color_b.py script by Robert L. Campbell, Copyright (c) 2004. Used with permission.
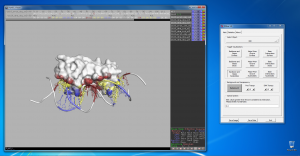 PDIviz is a plugin for the PyMOL molecular visualization system that is specifically designed to visualize protein-DNA interfaces in three dimensions. PDIviz detects the interface of protein-DNA complexes by calculating the buried surface area directly in PyMOL. The plugin provides a total of nine distinct 3D visualization modes to highlight interactions with DNA bases and backbone, major and minor groove, and with atoms of different pharmacophoric type.
PDIviz is a plugin for the PyMOL molecular visualization system that is specifically designed to visualize protein-DNA interfaces in three dimensions. PDIviz detects the interface of protein-DNA complexes by calculating the buried surface area directly in PyMOL. The plugin provides a total of nine distinct 3D visualization modes to highlight interactions with DNA bases and backbone, major and minor groove, and with atoms of different pharmacophoric type.
Installation notes:
- Unzip the downloaded plugin file and install with PyMOL’s Plugin Manager
- A licensed version of PyMOL is needed (incentive or educational)
- XQuartz is required on Mac OS: https://www.xquartz.org/ (https://www NULL.xquartz NULL.org/)
Usage:
1. Load a protein-DNA complex in PyMOL, e.g. execute the command: fetch 1bl0
2. Open the plugin from the Legacy Plugins menu
3. Click on one of the nine visualization buttons
Legacy version for PyMOL 1.x:
Download: PDIviz123 (v1.2.3 [2018-03-12])
Contacts:
Original algorithm: Andreas Schueller <aschueller@bio.puc.cl>
Plugin implementation: Judemir Ribeiro <jribeiro@uc.cl>
Project leader: Francisco Melo <fmelo@bio.puc.cl>
How to cite:
Ribeiro J, Melo F, Schüller A. PDIviz: analysis and visualization of
protein-DNA binding interfaces. Bioinformatics. 2015 Aug 15;31(16):2751-3. doi:
10.1093/bioinformatics/btv203 (https://academic NULL.oup NULL.com/bioinformatics/article-lookup/doi/10 NULL.1093/bioinformatics/btv203). Epub 2015 Apr 16. PubMed PMID: 25886981 (https://www NULL.ncbi NULL.nlm NULL.nih NULL.gov/pubmed/25886981); PubMed
Central PMCID: PMC4528634 (https://www NULL.ncbi NULL.nlm NULL.nih NULL.gov/pmc/articles/PMC4528634/).

 (http://www
(http://www