Research
Modeling of Protein-DNA Interactions by Adaptive Optimization
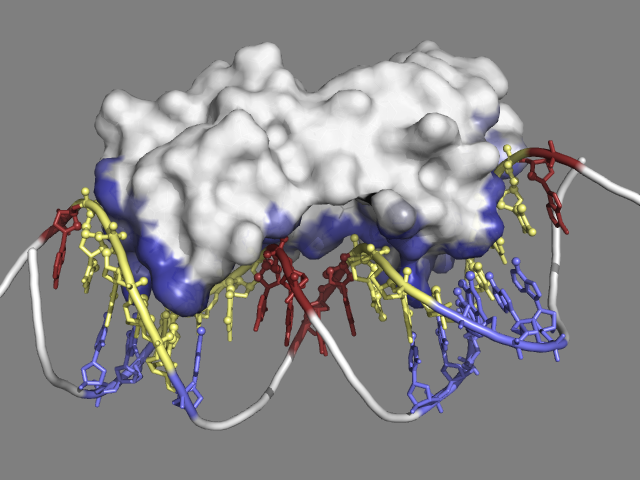
Multiple antibiotics resistance activator (MarA) bound to DNA (PDB code: 1bl0)
Protein-DNA interactions play pivotal roles in many biological processes such as DNA transcription, replication and repair. A key function is the regulation of genetic processes by specific interaction of transcription factors (TF) with DNA. MarA (multiple antibiotics resistance activator) is a member of the AraC/XylS family of TFs found in a variety of medically important bacteria, particularly the Enterobacteriaceae. MarA functions as a transcriptional activator which induces the expression of several genes providing different degrees of resistance to antibiotics.
Our research aims at the design of peptide inhibitors to prevent the interaction of MarA with its binding sites in the promoter region of its target genes. We analyse protein-DNA binding interfaces extracted from the protein-DNA interface database (PDIdb) to better understand the molecular basis of sequence recognition. This information then provides the starting point for the design of small peptides by adaptive optimization. Starting from an initial design an optimization algorithm improves the peptide sequence and thus the binding properties in an iterative fashion. The results of these in silico studies are experimentally validated by biochemical binding assays. Successful peptides will provide valuable feedback to the in silico design of peptides as tool compounds for the modeling of protein-DNA interactions.
This research is supported by a FONDECYT postdoctoral research grant (N° 3110009), FONDECYT (N° 1110400) and ICM (N° P09-016-F).
Full-atom Structure-based Prediction of Transcription Factor Binding Sites
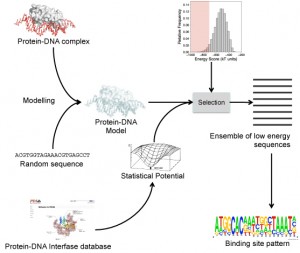
Flowchart for recovering known experimental TF binding sites
Protein-DNA binding is of paramount importance since it is involved in cellular processes such as gene expression and cell division. Since the first DNA-protein structure complex was solved at atomic resolution, our knowledge about how the recognition is carried out has increased notably.
Two main approaches to predict protein binding sites in the DNA have been reported: 1) Sequence-based methods, that use patterns or profiles coming from sequence alignments (e.g. consensus sequences, WebLogos) and 2) Structure-based methods, that use structural information coming from protein-DNA complexes solved by X-ray crystallography or NMR. The sequence-based methods are the most popular because of the simplicity in their use and implementation. However, these methods are not very accurate, exhibiting poor sensitivity/specificity trade offs.
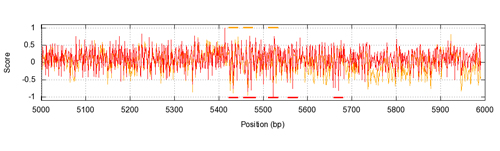
Prediction profile of TF binding sites at DNA sequence level using structural information
In our laboratory, we have been working on developing new statistical potentials that describe the protein-DNA interactions at atomic level, which can be used to estimate the stability of proteins-DNA complexes from the atomic coordinates of their 3D structures. We have combined the use of these potentials together with software for the 3D modelling of protein-DNA complexes, which has allowed us to recover in a large degree the known experimental binding sites for several transcription factors (TF). This approach has also allowed us to predict TF binding sites at DNA sequence level by using information available in the protein-DNA complex three-dimensional structures.
This research is supported by grants from FONDECYT (1110400 and 3120007) and ICM (P09-016-F).
Comparative Modeling of DNA and Protein-DNA complexes
The protein-DNA interactions knowledge is useful to increase the biological understanding of how is the regulation of genes controlled. Given that the Comparative Modeling of proteins has been a useful tool to predict protein structures and complexes when there are no real structures available, we want to develop a protocol to extend the Comparative Modeling to molecules that are not necessarily proteins. In this case, the DNA molecules are our target.
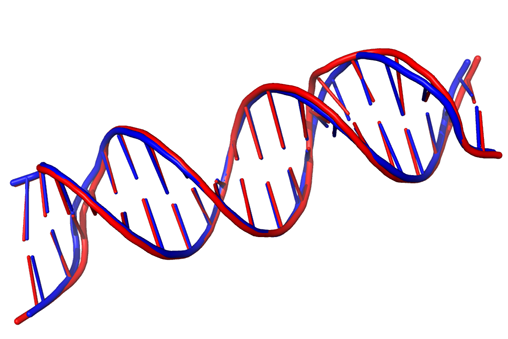
Comparative Modeling of 2V3L using homology derived restraints. (Model in Red, Template in Blue). Atomic RMSD = 0.97.
To accurately model the 3D structure of DNA and protein-DNA complexes by means of Comparative Modeling, models are being built using MODELLER. To facilitate this, statistical and stereo-chemical restraints applicable to nucleic acid residues and nucleic acid-amino acid interactions have been derived using real structures from the PDB. This set of values is being added as restraints to the MODELLER optimizer depending of the type of DNA we want to generate. To test the accuracy of the generated models, they are benchmarked against known 3D structures of DNA and protein-DNA complexes.
We suggest that these models will help make predictions on the DNA binding specificity of proteins. The obtained predictions will be tested in the laboratory.
This research is supported by a grant from FONDECYT (1110400).
DNA Damage Recognition
There is a consensus among the scientific community that aging is the result of accumulative and stochastic damage to biological macromolecules. One of these macromolecules is DNA. Proteins that recognize damaged DNA (to trigger repair pathways) can recognize different types of lesions without chemical similarity between them. A single lesion can be recognized by different proteins and pathways. It is currently unknown how damage detection proteins (DDPs) recognize different adducts embedded in the genome and how different proteins detect the same adduct.
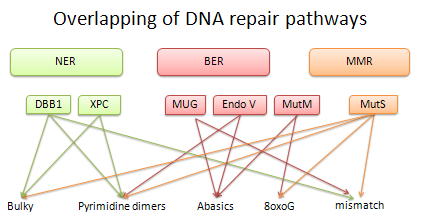
Overlap in DNA-damage detection of proteins belonging to Nucleotide Excision Repair (NER) Base Excision Repair (BER) y Mismatch Repair (MMR)
To address these questions our research is focused on structural bioinformatics and biophysical experimentation. We have built a structural database of protein-damaged-DNA complexes and isolated damaged-DNA. We have found similarities between DDPs and between different types of lesions. For us, the structural bioinformatics study provides a starting point to tests our findings experimentally.
This research is supported by grants from FONDECYT (1110400) and ICM (P09-016-F).
Study of Sequence-Structure-Function Relationships in Peptides and Oligonucleotides
Protein fragments have been, for a long time, an important research subject in bioinformatics, with applications in protein homology modeling, secondary and tertiary structure prediction, study of sequence-structure relationships and drug design. In this study, we developed a repository containing thousands of protein fragments clustered by structural similarity. These fragments are extracted from different protein datasets, which include a large number of peptides from a non-redundant set of monomeric and globular proteins (MGP), a non-redundant dataset of protein-DNA complex interfaces (PDIdb), a database of protein-DNA complexes (PDC), a non-redundant set of trans-membrane (TM) proteins, and a non – redundant set of RNA structures, among others.
The database also includes information like sequence conservation, secondary structure and solvent accessibility surface area were calculated for every fragment in the context of the native structure.

Sample clusters from the FragProt database: peptide fragment cluster (left), cluster of RNA structures (right)
Finally, a structure-based search method was implemented using the Ultrafast Shape Recognition (USR) algorithm, allowing the user to provide a fragment in PDB format as a query to search in the database. These improvements, in addition to other features like different Jmol visualizations and dynamic searches, make this database a useful tool to explore the growing universe of peptide conformations and to study the sequence-structure-function relationships in proteins.
This research is supported by grants from FONDECYT (1110400) and ICM (P09-016-F).
Next-Sequencing Technologies Analysis (RNA-seq, Metagenomics, Exome-seq, ChIP-seq, Small Genome Assembly)
The advent and integration of high-throughput ‘omics technologies are becoming instrumental to assist fundamental explorations of the systems biology of organisms. In particular, these technologies now provide unique opportunities for global molecular investigations. For example, studies of the transcriptome of different species and/or developmental stages provide insights into aspects of gene expression, regulation and function, which is a major step to understanding their biology. The amazing acceleration in biological inquiry enabled by the current next-generation sequencing instrumentation is clearly just beginning. These instruments will continue to evolve, and new platforms just introduced, or under development, will have a continuing impact on biological and biomedical research for years to come.
Computational tools for the analysis of NGS data
The aim of this research is the ability to deliver services related to the analysis of vast amounts of information, committed both by the different sequencing platforms (e.g. Illumina, SOLID) and the various experiments that can be performed with them (e.g. RNA-seq, ChIP-seq). Considering the large and diverse range of experiments possible in the hundreds of species models, our efforts are focused on two areas. One is the core design that aims to address common needs among various experiments (e.g. filtering, mapping, counting). The other area is customized design to the needs of the researcher. As a result of collaborations with other labs, we have already conducted analysis of small RNAs in gastrula stage, during the embryonic development of Xenopus tropicalis. Currently, we are working on the differential expression of transcripts for spinal cord regeneration in Xenopus laevis.
This research is supported by grants from ICM (P07/011-F, P09/016-F), BASAL PFB12/2007, and FONDECYT (11100348, 1110400).
Structure-Based Analysis of Affinity and Specificity Determinants in MHC-Peptide-TCR Complexes
The major histocompatibility complex (MHC) is a set of molecules that play a key role in the immune system. When a pathogen infects the cell, pathogenic proteins are processed by the antigen processing machinery into peptides and only a small fraction is loaded onto the MHC molecules. The peptide-MHC complexes (pMHC) are expressed on the cell surface and the immune response is elicited via T-cell receptor (TCR) binding.
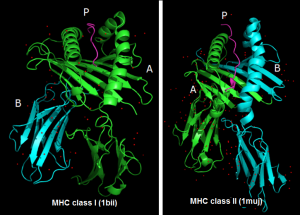
Examples of two curated structures present in the 3D-pMHC database
The understanding of the structural principles involved in the selection of specific antigenic peptides by the different MHC alleles and TCR/pMHC recognition is critical in drug and vaccine design. 3D-pMHC is a curated database of peptide-MHC Class I and II complexes containing structures of 367 complexes with a resolution <= 3.5 Å. To create the database, the complete Protein Data Bank (PDB, release Nov 2011) was screened with TopSearch, a fast structure comparison software tool. Afterwards, from each structure, the pMHC complex was extracted and the protein chains renamed in a standardized way. Structures lacking the peptide or peptides with missing residues, fusion proteins, MHCs bound to non-classical ligands and other unrelated proteins similar in structure to the MHCs were discarded from the database. Finally, an analysis of sequence redundancy was carried out, thus generating a database that contains only unique entries.
The aim of building the 3D-pMHC database is to enhance the understanding of the binding mechanism of the pMHC complexes, in order to obtain information with predictive value for future development of new drugs and vaccines.
This work is funded by grant P09/016-F from Iniciativa Científica Milenio (ICM).
Statistical and Conformational Analysis of Canonical and Non-Canonical Base Pairs in RNA Three-Dimensional Structures
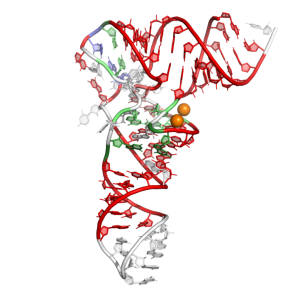
Base pair pattern of t-RNA PHE (PDB: 3L0U) colored by the PyMOL plugin
Classically, RNA molecules are given the title of biological information carrier used in the synthesis of proteins and ribosomal structural cores. Lately, new RNA functions are being discovered, such as their biological activity as regulators and catalysts, and their role in cell signaling. To better understand the function of RNA molecules, the study of their three-dimensional structures is required. Features like canonical and non-canonical base pairing and base stacking are essential driving forces that stabilize this molecule. The principal focus of this work is the statistical analysis of RNA base pairs identified from a reduced-redundancy set of RNAs by the RNAView software. Conformational and statistical analysis information can be used for RNA modeling purpose.
In order to have a better understanding of RNA structure, we are further developing a simple tool that facilitates base pair visualization. PyMOL is a well know molecular viewer used in structural investigation, but visualization of nucleic acids is currently limited in this software. For this reason we are developing a PyMOL plugin that applies a coloring scheme for base pairing implementing the nomenclature proposed by Leontis and Westhof.
This research is funded by grants from FONDECYT (1110400) and ICM (P09-016-F).

 (http://www
(http://www